Genetics of Oxygen Deprivation in Marine Mammals and Humans
Project Team
Team profile by members of the Learning From Whales project team
As oceans warm due to climate change, dissolved oxygen concentrations are declining. Many marine animals could suffer from this hypoxic stress, or stress due to insufficient oxygen levels. However, certain marine animals tolerate a lack of oxygen all the time: whales and dolphins. When these cetaceans dive to deep depths to forage for food, they hold their breaths the entire time. These charismatic organisms provide an excellent model to discover the basis of hypoxia tolerance, which could be leveraged to protect more sensitive species.
Understanding hypoxia tolerance has applications for human health as well. Oxygen deprivation in human tissues causes mass cell death in heart attacks and strokes; hypoxic conditions are also a driving force in cancer metastasis. Illuminating the genetic responses to hypoxia across species can improve our management of these human health issues.
Our team divided into two sub-teams to analyze hypoxia tolerance from different angles. Nikki, Samantha, and Giselle analyzed genetic variation between the two ecotypes using RAD-sequencing and sequencing of the mitochondrial control region. Larry, Raksha, and Elizabeth analyzed gene regulation data to characterize how different genetic pathways respond to hypoxia.
RAD-sequencing and Mitochondrial Control Region Sequencing

To investigate adaptations of marine mammals to hypoxia, we employed Restriction Site Associated DNA sequencing methods to assess genetic variability and estimate an effective population size of inshore and offshore bottlenose dolphins (Tursiops truncatus) in the Northwest Atlantic.
Analysis of 14,783 single nucleotide polymorphisms revealed at least three genetically differentiated populations. Our results suggest an inshore population along North Carolina’s Outer Banks (n=32), an offshore population off the continental shelf break from North Carolina to Jacksonville, Florida (n= 38), and a shelf population off Jacksonville, Florida (n=26).
Bayesian clustering showed significant admixture between the North Carolina and Jacksonville populations, providing potential evidence of historical or current gene flow. Our analyses provide fine-scale genetic resolution of bottlenose dolphin population differentiation in the Western North Atlantic.
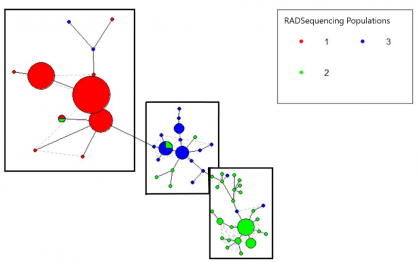
This data provides a baseline for continued research into comparing genetics of inshore and offshore ecotypes and exploring how they relate to differences in diving behavior and adaptations to hypoxia. Genome wide variation will provide a powerful dataset for understanding the genetic makeup underlying hypoxia tolerance in cetaceans.
Mitochondrial control regions were sequenced and then put into a haplotype network. The haplotype network revealed three populations, and these populations were compared with those determined from RAD-sequencing.
Molecular Interactions of Hypoxia
Larry, Raksha, and Elizabeth analyzed the relationship of hypoxia with stress, cell motility, and metabolism, respectively. These three physiological responses are critical to marine mammal diving behavior and survival. Using a systems-level approach, gene sets from each of the three pathways were downloaded from online databases, and interactions were investigated through gene network analysis. An experimental approach was also used to analyze hypoxia’s interaction with glucocorticoid, the molecular stress response. Addition of cortisol to Cuvier’s beaked whale cells (Ziphius cavirostris) suppressed the hypoxia response. Furthermore, Raksha used phylogenetic and evolutionary analysis to discover that there is evidence for the coupling of the hypoxia and cell motility pathways early in eukaryotic evolution.
Each of these molecular responses was found to be closely linked to hypoxia, and key genes mediating the interaction between hypoxia and stress, cell motility, and metabolism were identified. By identifying core genes of these responses to hypoxia and how different species are genetically adapted to hypoxia, we can improve conservation efforts to help marine mammals and other organisms survive in increasingly stressful, hypoxic oceans.
Learning from Whales: Identifying Key Genes in Genetic Pathway Responses to Low Oxygen
Poster by Larry Zheng, Elizabeth Bock and Raksha Doddabele
Using Genetics to Understand Population Structure of Inshore and Offshore Bottlenose Dolphins (Tursiops truncatus)
Poster by Samantha Townsend, Nikki Shintaku, Nicola Quick, Tom Schultz, Jason Somarelli, Jillian Wisse, Ashley Blawas, Elizabeth Bock, Raksha Doddabele, Giselle Wang and Larry Zheng


